Research
RESEARCH PROJECT
Glycobiology in the Cytosol and Drug Discovery Based on Glycoprotein-Specific Ubiquitin Ligases
Proteins are glycosylated in the endoplasmic reticulum (ER) or Golgi apparatus. These membrane-bound organelles transport glycoproteins to the extracellular matrix or to other organelles. Because of this transport system, glycoproteins were not thought to be present in the cytosol. However, recent studies have found that glycoproteins transiently appear in the cytosol after bacterial infection, organelle damage, or glycoprotein misfolding.
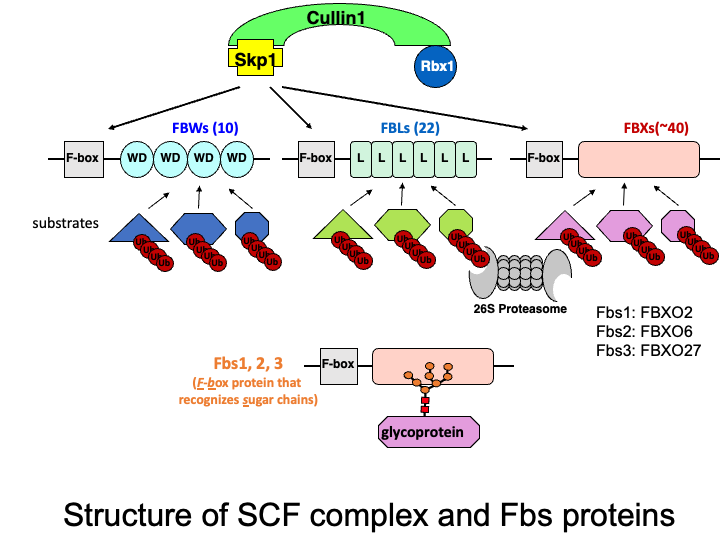
The Skp1-Cullin1-F-box protein containing complex (or SCF complex) is an E3 ubiquitin ligase complex that ubiquitinates proteins to direct them toward proteasomal degradation. F-box proteins are substrate-recognition subunits of the complex, and we previously identified three cytosolic F-box proteins (F-box protein recognizing sugar chains: Fbs1, Fbs2, and Fbs3) that recognize N-linked glycans. Currently we are studying the function of these F-box proteins and identifying target glycoproteins in the cytosol.
☆ Glycoprotein-specific ubiquitin ligases
The ubiquitin-proteasome system is responsible for selective proteolysis in eukaryotes. Ubiquitination is catalyzed by three enzymes-E1, E2, and E3, with the E3 enzyme providing target selectivity. The human genome encodes more than 600 E3 enzymes, and the SCF complex consists of three invariant components (Skp1, Cullin1, and Rbx1), and a variable F-box protein that functions as a substrate recognition protein. Among the 70 F-box proteins in humans, we have been characterizing three F-box proteins that recognize glycans.
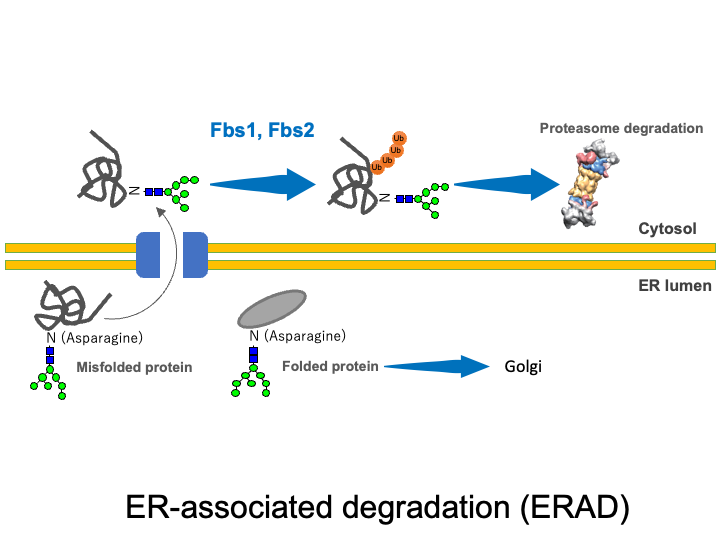
☆ ER-associated degradation (ERAD) pathway
Extracellular proteins such as secretory proteins and membrane proteins are translated in the rough endoplasmic reticulum (ER) and then enter the lumen of the ER, where sugar chains are attached. In the ER, protein folding is carried out with the help of ER chaperones. Proteins that were not folded correctly as well as excess protein subunits are returned from the ER to the cytosol (retrograde transport) where they are degraded by the UPS. This quality control mechanism is known as the ERAD system. SCF complexes containing Fbs1 and Fbs2 are thought to participate in ERAD by recognizing the innermost N-glycans in glycoproteins (the GlcNAc-GlcNAc structure commonly present at the base of N-glycans) that are exposed in denatured proteins or misfolded proteins.
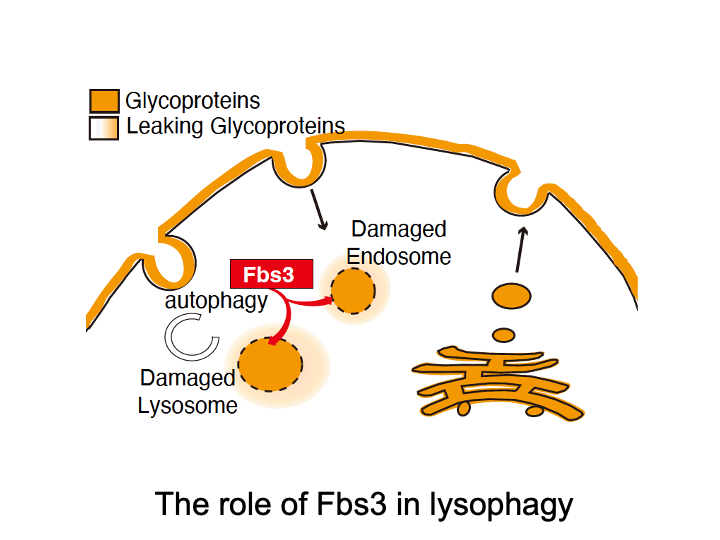
☆ Lysophagy
Fbs3 differs from other F-box proteins because it is myristoylated and localizes to intracellular organelle membranes. When the lysosomal membrane is damaged by bacterial infection or by microparticles taken up from outside the cell, a glycoprotein called LAMP2 becomes exposed to the cytoplasm. SCF complexes containing Fbs3 quickly accumulate around LAMP2 and ubiquitinate many proteins on the damaged lysosome. This induces autophagy and degradation of the damaged lysosome, a process called lysophagy.
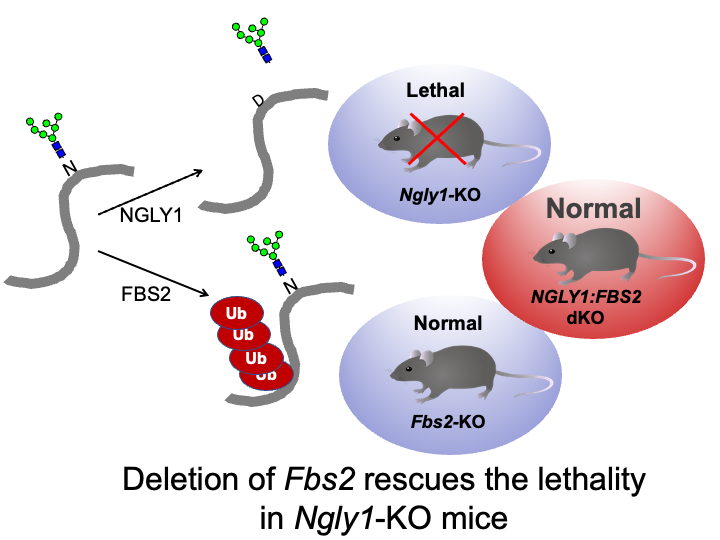
☆ NGLY1 deficiency
Besides glycoprotein-specific ubiquitin ligases, glycosidases that remove sugar chains from proteins are also found in the cytoplasm. N-glycanase (NGLY1) deglycosylates various substrates before they are degraded by the proteasome in the ERAD pathway. Recently, a serious systemic disease called NGLY1 deficiency, an ultra-rare disease with mutations in NGLY1 has been found via exome analysis. Dr. Tadashi Suzuki at RIKEN, who studies NGLY1, reported that Ngly1-KO mice (B6 strain) were embryonic lethal. By chance, we found that mice that are knocked out for both Ngly1 and Fbs2 are born and grow normally. In addition, cell death is observed when Fbs2 is expressed in Ngly1-KO cells, suggesting that Fbs2 is deleterious in NGLY1-KO cells and animals. Based on these findings, we believe that development of inhibitors of Fbs2 may be a promising strategy for treating NGLY1 deficiency and we have currently started screening for these types of drugs with Dr. Suzuki. We are also working to elucidate how Fbs2 causes cytotoxicity.
Intracellular Dynamics of the Proteasome
The proteasome is a huge proteolytic enzyme complex assembled of 33 subunits, which was discovered by Dr. Keiji Tanaka, the supervisor of our lab. Dr. Yasushi Saeki (currently a professor at the Institute of Medical Science, The University of Tokyo) analyzed intracellular dynamics of the proteasome by attaching a fluorescent protein to a proteasome subunit. Currently, Dr. Endo in this lab has been analyzing dynamic changes in the proteasome upon various stresses, and unraveling exciting phenomena.
Comprehensive Understanding of the Ubiquitin-Proteasome System
Ubiquitin is a small protein consisting of 76 amino acids that forms various chains through covalent bonds using seven Lys residues and the N-terminal Met. Different chains have different functions. For example, K48 chains are known to direct proteins to degradation by the proteasome. Dr. Tsuchiya and his colleagues have clarified the shuttling mechanism by which K48-modified substrates are transported to the proteasome. Currently, in collaboration with Dr. Saeki, Dr. Tsuchiya is comprehensively analyzing the shuttle molecules that transport ubiquitinated substrates to the proteasome and Dr. Tomita is analyzing molecular mechanisms involved in quality control for protein complexes.
Functional analysis of proteasome mutant mice
Disruptions of proteasome function are thought to cause a variety of diseases including neurodegenerative disorders. However, since proteasome function is essential for cell survival, pathophysiological analyses of animal models of proteasome dysfunction are lacking. To address this issue, our laboratory has created a mouse model of systemic proteasome dysfunction using a heterozygous mutation in the proteasome subunit, PSMD12, found in a patient with a developmental disorder. Currently, in collaboration with Dr. Saeki, Dr. Tsuchiya and Dr. Yonekawa are analyzing proteasome functions in this mouse model.

