
HOME > Project > Laboratory of Protein Metabolism
Laboratory of Protein Metabolism

Laboratory Head
Yasushi Saeki
Laboratory of Protein Metabolism
Brochure 2018_PDF ![]() (349.8KB)
(349.8KB)
Backgrounds
Protein Metabolism:
The Ubiquitin Proteasome System
All proteins of the cell continually are recycled with individually distinct life-span, which is fundamentally important to keep healthy cellular activities in eukaryotes. The proteasome, in collaboration with the ubiquitin system used for choice of target proteins, selectively degrades unnecessary proteins that must be eliminated from the cells. Indeed, the ubiquitin-proteasome system (UPS) plays a pivotal role in the control of a diverse array of basic cellular functions by catalyzing a number of reactions timely, rapidly, and irreversibly. The missions of our laboratory aim to elucidate the molecular mechanisms of the UPS and to integrate it into physiology and pathology.
Research Projects The Proteasome
1. Structure and Assembly of Proteasomes
The 26S proteasome is a highly organized and sophisticated proteolytic nanomachine that degrades ubiquitylated proteins in an ATP-dependent fashion. (Fig. 1). One longstanding question is how the complex structure of the proteasome is organized with high fidelity. During the past decade, we have succeeded in structural analysis of the 20S and 26S proteasomes. Further, we identified approximately 10 proteasome-dedicated assembling chaperones (i.e., CP and RP chaperones) that assist in the efficient formation of 20S and 26S proteasomes. Of them, we have determined the tertiary structures of almost all CP and RP chaperones, displaying the mechanisms underlying proteasome assembly at the atomic levels. Based on these findings, we have established a model in which multiple dedicated chaperones govern proteasome assembly.
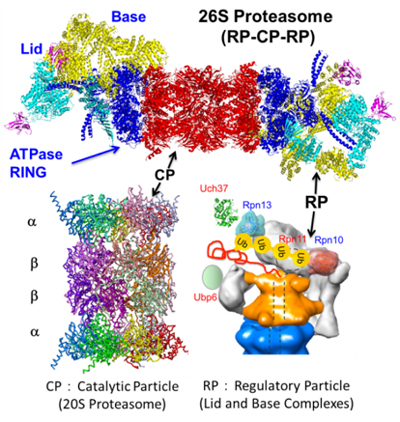
The 26S proteasome consisting of a 20S core/catalytic particle (CP) and one or two 19S regulatory particles (RP). The CP (alias 20S proteasome) is composed of four heptameric rings, which are made up of seven structurally related α and β subunits, displaying an α1-7β 1-7β1-7α1-7 organization. The RP recognizes ubiquitylated proteins, deubiquitylates for recycling of ubiquitin, and then unfolds and translocates them into the interior of the CP for degradation.
2. Roles of Specialized Proteasomes in Cell-mediated Immunity
The proteasome has acquired diversity of the catalytic β subunits, which have evolved during the acquisition of adaptive immunity. To date, we have discovered the vertebrate-specific alternative 20S CP, which we named the "immunoproteasome" and the “thymoproteasome” (Fig. 2). Whereas the immunoproteasome plays a specialized role as a professional antigen-processing enzyme in cell-mediated immunity, the thymoproteasome is involved in the development of CD8+T cells in thymus; i.e., it has a key role in the generation of MHC class I-restricted CD8+T cell repertoire during thymic selection called positive selection.
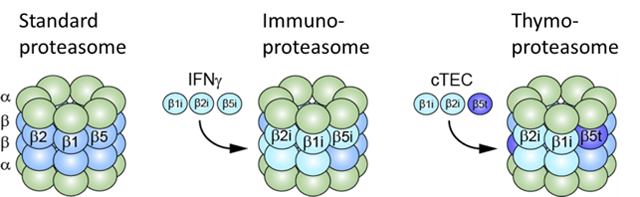
The immunoproteasome has catalytic subunits β1i, β2i, and β5i replacing β1, β2, and β5 and enhances production of MHC-I ligands. The thymoproteasome contains thymus-specific subunit β5t in place of β5 or β5i and plays a pivotal role in positive selection of CD8+ T cells.
3. Spatio-temporal Dynamisms of the Proteasome
Little is known about the molecular dynamics of the proteasomes in living cells. We measured the absolute concentration, dynamics, and complex formation of the proteasome by quantitative live-cell imaging and quantitative proteomics analyses. We found that the 26S proteasome is a highly mobile complex and enriches in the nucleus. The 26S proteasome appears to complete its assembly process in the cytoplasm and then translocates as a holoenzyme into the nucleus. Furthermore, we also found that the proteasome dynamically changes its subcellular localization and its cofactors under various stresses. This might be a novel cellular response for adapting to stress by regulating protein homeostasis.

