HOME > Topics2019 > 15 April 2019
15 April 2019
Daisuke Yamane (Viral Infectious Diseases Project) published a paper on “Basal Expression of Interferon Regulatory Factor 1 Drives Intrinsic Hepatocyte Resistance to Multiple RNA Viruses” in Nature Microbiology
Discovery of a Previously Unappreciated Layer of Hepatocyte Innate Immunity against Pathogenic RNA Viruses
Summary
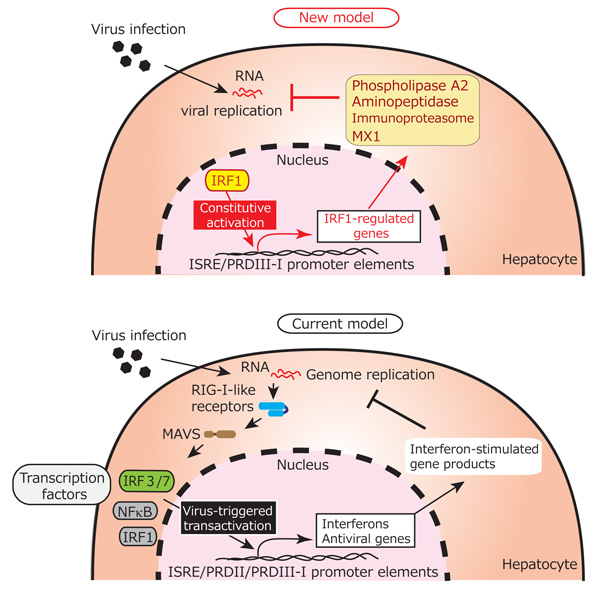
It has long been recognized that hepatocytes possess a potent innate immune system, which immediately eliminates infection when primary cultures are challenged with liver tropic RNA viruses. A collaborative research conducted at Tokyo Metropolitan Institute of Medical Science and the University of North Carolina at Chapel Hill has discovered a mechanism by which hepatocytes’ intrinsic resistance to diverse RNA viruses is regulated.
- <Title of the paper>
- Basal expression of interferon regulatory factor 1 drives intrinsic hepatocyte resistance to multiple RNA viruses
- <Journal>
- Nature Microbiology
https://www.nature.com/articles/s41564-019-0425-6
DOI: 10.1038/s41564-019-0425-6
Details
A research team led by Dr. Daisuke Yamane at the Infectious Diseases Project in Tokyo Metropolitan Institute of Medical Science and Professor Stanley M. Lemon at the University of North Carolina at Chapel Hill has discovered that the intrinsic resistance of hepatocytes against RNA viruses is primarily driven by basally expressed interferon regulatory factor 1 (IRF1).
Primary hepatocytes are refractory to virus infections unlike the permissive hepatoma cell lines (e.g. Huh-7 and HepG2 and their derivatives) which express defective antiviral signaling components, but the cell-intrinsic mechanism has remained poorly characterized due to difficulties in manipulating gene expression in primary hepatocyte cultures. The research team took advantage of a unique cell line originally derived from immortalized adult human hepatocytes. PH5CH8 cells are unique in that they retain expression profiles of intact antiviral components similar to hepatocytes in vivo. By depleting a series of host antiviral components in PH5CH8 cells, the research team mapped detailed host signaling pathways that restrict diverse RNA viruses and found a pivotal protection mechanism driven by constitutively expressed IRF1 to regulate basal antiviral state of hepatocytes. This was surprising, as antiviral responses in hepatocytes have thought to be governed by the virus-triggered responses, which is induced in response to cellular sensing of pathogen-associated molecular patterns by the sensor proteins, such as RIG-I-like receptors or Toll-like receptors, to initiate host interferon responses. However, the data presented in this work demonstrate that constitutively expressed IRF1 is predominant in restricting virus infections through regulating basal expression of a suite of antiviral genes, including genes with phospholipase A2 activity, aminopeptidases, components of immunoproteasome, and apolipoproteins etc., many of them were previously not known to have antiviral activity.
Further studies to elucidate the mechanism of antiviral action elicited by each of the IRF1 effector genes would allow us to uncover fresh strategies to treat pathogenic virus infections by mimicking host antiviral responses.
Reference figure