Meet our scientists!
 Back
Back
We know intuitively that we have internal circadian clocks by our tendency to wake up in the mornings and go to sleep at night. This tendency continues even when we are in constant darkness where we shouldn’t be able to tell whether it is day or night. But circadian clocks control many more behaviors than just sleep patterns. Clocks are found in plants, animals, and bacteria, and they govern almost all aspects of biology. The most important and intriguing characteristic of clocks is their oscillation. While we understand the physics behind oscillations of pendulums and springs, biological processes tend to approach a state of equilibrium where they become stable and don’t oscillate. So, the study of biological clocks is important for biological and medical reasons, but also simply as a fascinating intellectual mechanism to understand. Currently, we’ve identified some aspects of circadian clocks. Two transcription factors, CLOCK and BMAL1, work together and bind to DNA sequences called E-boxes to activate transcription of genes, including PER and CRY. PER and CRY then inhibit CLOCK and BMAL1 activity, creating a rhythmic oscillation that is thought to be a core clock mechanism. However, this model is too simple and many questions still remain. Yuta Otobe and colleagues in the Circadian Clock Project at TMIMS are working on completely elucidating circadian clock mechanisms. Recently they found that phosphorylation affects DNA binding and PER-dependent inhibition of CLOCK and BMAL1 to regulate the function and frequency of the circadian clock (PNAS (2024), 121:23, e2316858121). We spoke to Dr. Otobe about his work.

What first sparked your interest in biology?
When I was a child, there was a television show on reptiles hosted by Dr. Shouichi Sengoku. Dr. Sengoku loved reptiles and influenced many children, including me, to become fascinated by them. In my kindergarten, we raised a turtle, and I remember how worried I was when he escaped one day and how relieved I was when I found him six months later hibernating in the cold. Those early experiences made me love animals and made me want to become a researcher to understand them better. My interest in circadian clocks came later when I was a student because I had a really hard time organizing my sleep/wake routines and other daily schedules. I wanted to know why my schedule kept getting later. I thought that if I could understand circadian rhythms, I could understand why it was so difficult for me to organize my daily rhythms.
How did you get involved in this current research?
I really have to acknowledge Mr. Shunsuke Ito, the third author of our paper. He was a former student who laid the foundations for this work. Cultured cells have circadian rhythms that we can measure through activity of reporter genes. Shunsuke knocked out CLOCK and BMAL1 in cultured cells and showed that these knockouts abolished circadian rhythms. He then added back CLOCK and BMAL1 and showed that rhythms were restored. The importance of this work was that it allowed us to now add back CLOCK and BMAL1 with different mutations to see how mutations affected rhythms.
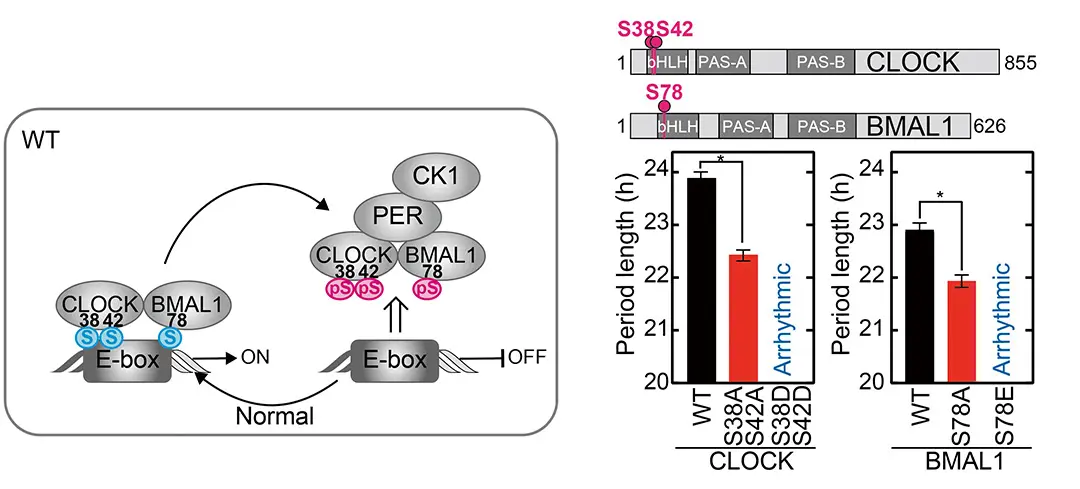
We were interested in phosphorylation, which is a mechanism that modifies proteins to alter their function, so I inserted various mutations in phosphorylation sites in CLOCK and BMAL1. To summarize our results, I found that preventing phosphorylation of serine 38 and 42 in CLOCK and serine 78 in BMAL1 by mutating these serines to alanine, shortened the circadian cycle. In the converse experiment, mimicking phosphorylation at these sites, by mutating them to glutamate, abolished rhythms. Previously, we’d found that phosphorylation of these sites inhibits CLOCK and BMAL1 from binding to DNA to activate transcription. In this current study, we showed that phosphorylation of these sites further regulates the period of the circadian clock.
How do your findings help us understand the circadian clock better?
We formed an international collaboration with Dr. Jae Kyoung Kim’s group in Korea who helped us model the interactions between CLOCK, BMAL1, DNA, CRY, and PER. A previous model, called the Kim-Forger model, has been used to model oscillations of the circadian clock where CLOCK and BMAL1 activate transcription of PER and CRY which subsequently inhibit CLOCK and BMAL1 in a cyclic manner. In the Kim-Forger model, PER inhibits transcription by sequestering CLOCK and BMAL1 away from E-box DNA, while CRY inhibits transcription by directly binding to and inhibiting BMAL1. However, more recent studies have identified a different mechanism by which PER inhibits CLOCK-BMAL1-dependent transcription. In PER-dependent displacement, PER recruits the kinase, casein kinase I, which phosphorylates CLOCK to remove the CLOCK-BMAL1 complex from the E-box. We incorporated this new mechanism into the Kim-Forger model and conducted simulation studies to determine how our phosphorylation site mutations should affect the circadian period. While our serine-to-alanine mutations should decrease the dissociation constant of CRY-CLOCK-BMAL1 from DNA to lengthen the period, they were also able to decrease the dissociation constants of both PER-CLOCK-BMAL1 and PER-CRY-CLOCK-BMAL1 from DNA to shorten the circadian period in agreement with our experimental results. This indicates that our serine-to-alanine mutation may disrupt the PER-dependent displacement of the CLOCK-BMAL1 complex from DNA. Currently, I believe that serine 38 and 42 on CLOCK and serine 78 on BMAL1 are phosphorylated by a kinase, casein kinase I, which both decreases binding of CLOCK and BMAL1 to E-box DNA, and also affects the ability of PER to displace CLOCK and BMAL1 from E-box DNA. This mechanism is important, at least in fine-tuning the clock frequency.
Wonderful work! What are your plans for the future?
I’ve got many ideas I’d like to pursue for the future!
In the near future, I’d like to continue my current work of better understanding the circadian clock. This year, the institute obtained a next-generation mass spectrometer that can analyze 100,000 proteins at once. I’d like to comprehensively analyze protein changes and fluctuations at different time points in different mutants in order to develop more accurate models of clock function. Also, although our current work depends on feedback loops (PER and CRY) affecting transcription (CLOCK and BMAL1), this is likely only part of the circadian clock mechanism. We currently have solid evidence demonstrating the existence of a transcription- and translation-independent clock oscillator. I’d like to continue this work to identify the true core clock oscillator. Another idea I have is to try to reconstitute the circadian clock in vitro using cell extracts or purified CLOCK, CRY, BMAL1, PER, and CKI proteins.
Finally, in the far future, after I’ve obtained my own lab, I’d like to devote part of my time to studying reptiles or some other animal that might draw the interest of children. Similar to how Dr. Sengoku inspired me to pursue a career as a research scientist, I’d like to contribute in some way toward teaching children the joys of studying animals.
Interviewed by Jun Horiuch