
HOME > Project > Laboratory of Protein Metabolism
Laboratory of Protein Metabolism

Laboratory Head
Yasushi Saeki
Laboratory of Protein Metabolism
Brochure 2018_PDF ![]() (349.8KB)
(349.8KB)
Backgrounds
Protein Metabolism: The Ubiquitin Proteasome System
The Ubiquitin System
1. Developing Methods to Decipher the Ubiquitin Code

Hikaru Tsuchiya
Ubiquitylation is involved in numerous important cellular processes such as proteasomal degradation, DNA repair, protein sorting, and signal transduction. The ubiquitin function is relied on eight structurally distinct ubiquitin chains of different lengths, but our knowledge of the relationship between their topology and functional outcomes is still insufficient. To understand the ubiquitin code, it is essential to develop new methods for analyzing linkage types, chain lengths, and complexity of ubiquitylation.
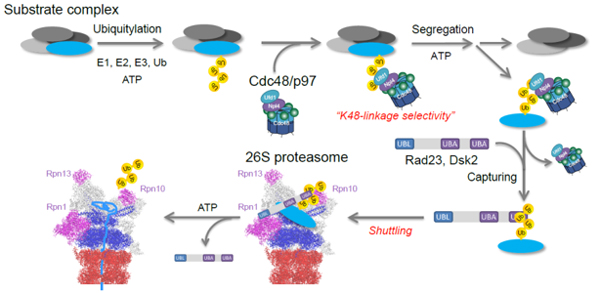
Proteasomal degradation is mainly regulated by indirect substrate sorting pathway by the ubiquitin-selective chaperone Cdc48/p97 and the shuttling factors Rad23 and Dsk2.
We have developed a highly sensitive MS/MS-based quantification method for ubiquitin chains. The method allows us to analyze linkage-type selectivity of ubiquitin decoder proteins at endogenous experimental setting. We recently identified the main pathway targeting the K48-linked ubiquitylated substrates for proteasomal degradation (Fig. 3). We are further analyzing the decoder proteins throughout the ubiquitin-mediated pathways to reveal the ubiquitin network.
2. Roles of Novel Ubiquitin Codes
Using quantitative mass spectrometry, we recently identified multiple chemical modifications of ubiquitin itself. Acetylation of ubiquitin inhibits the elongation of particular ubiquitin chains, whereas phosphorylation of S65 ubiquitin stimulates ubiquitin chain synthesis for mitophagy. More recently, we identified more complexed ubiquitin chains branched at K48 and K63. The K48/K63 branched chains act as a unique coding signal that specifically affects recognition by downstream reader proteins to enhance NF- κB signaling. These novel ubiquitin codes greatly expand ubiquitin functions. We further explore additional roles and regulatory mechanisms of these novel ubiquitin codes.

Fumiaki Ohtake
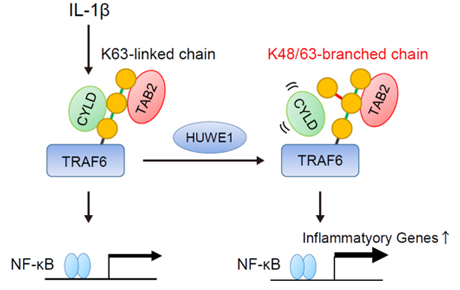
Upon IL-1β stimuli, two ubiquitin ligases, TRAF6 and HUWE1 cooperatively assemble K48/K63 branched chains to amplify NF-κB signaling.
On-going Projects
- Regulations of the proteasome to adapt to environmental stress
- Exploring proteasome cofactors as therapeutic targets
- Generation of mouse models of proteasome-related diseases
- Developing methods to analyze ubiquitin chain architecture
- Roles of the branched ubiquitin chains in protein degradation and signaling

