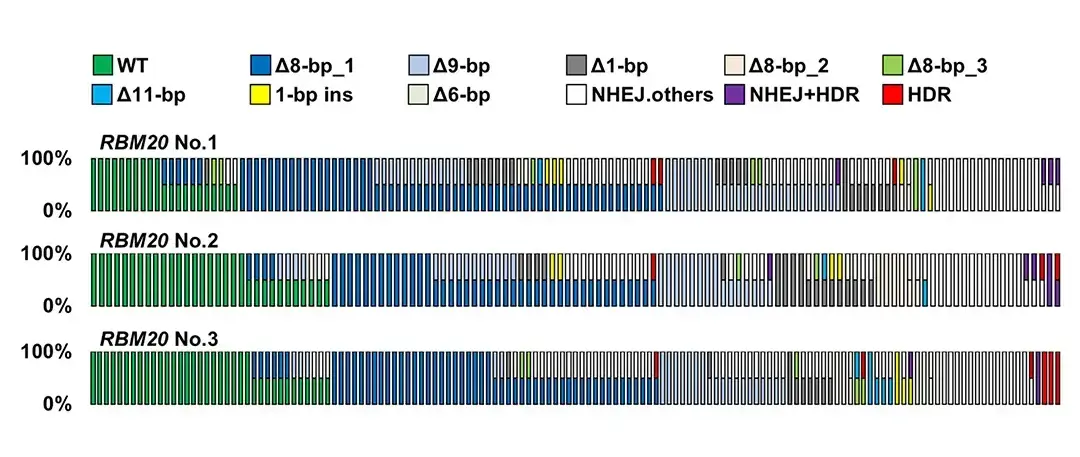
Genome editing in human iPS cells is important for the development of therapies for genetic diseases. However, genome editing with CRISPR-Cas9 is known to induce random insertions and deletions in addition to the desired editing. Since human cells have two sets of chromosomes, a genome-edited cell population will contain cells with various combinations of these sequences. Understanding the combinations and frequencies of various gene sequences generated by genome editing is important for estimating the therapeutic effects of editing. However, iPS cells are prone to cell death due to single cell isolation, and it has been difficult to establish and analyze a large number of clones using conventional single cell cloning methods.
Gou Takahashi (then Staff Scientist, now Affiliated Scientist, at Tokyo University of Agriculture), Minato Maeda (then Graduate Student) and Kayoko Shinozaki (Graduate Student) and Yuichiro Miyaoka (Project Leader) has succeeded in accurately isolating and analyzing over 1,000 genome-edited iPS cell clones by combining the Matrigel domes culture method, which forms cell clumps from single genome-edited iPS cells, and the CELL HANDLERTM (Yamaha Motor Co., Ltd.), which enables cell picking and accurate automated cell dispensing by an image recognition system. The analysis revealed frequent occurrence of identical genome editing in two target sequences in iPS cell. For example, a 1-bp insertion or an 8-bp deletion was respectively induced in both target sequences on homologous chromosomes in an iPS cell. This finding is a new insight into genome editing in human iPS cells and is expected to contribute to the development of treatments for genetic diseases.