HOME > Topics2018 > 6 February 2018
6 February 2018
Hikaru Tsuchiya, PhD, (Laboratory of Protein Metabolism) published a paper on “A novel versatile method for assessing chain length of substrate-attached polyubiquitin chains” in Nature Communications.
A novel versatile method for assessing chain length of substrate-attached polyubiquitin chains
Summary
Protein ubiquitylation regulates diverse cellular processes via distinct ubiquitin chain architectures assembled from eight different ubiquitin linkages and chains of various lengths. Despite the fundamental importance of chain length, the lack of a comprehensive method for measuring this property has limited our knowledge of its physiological relevance and regulatory mechanisms. Scientists at the Tokyo Metropolitan Institute of Medical Science and the Tokyo Institute of Technology developed a method for determining the chain length of ubiquitylated substrates. Using this method, we found that most endogenous ubiquitylated substrates in yeast soluble lysate are attached to chains with up to seven ubiquitin molecules. Unexpectedly, inactivation of the ubiquitin-selective chaperone Cdc48, rather than the proteasome, caused a dramatic increase in chain lengths on substrate proteins, suggesting that Cdc48 antagonizes the ubiquitylation machinery and terminates chain elongation by substrate extraction.
- <Title of the paper>
- Ub-ProT reveals global length and composition of protein ubiquitylation in cells
- <Journal>
- Nature Communications, 6;9(1):524.
doi: 10.1038/s41467-018-02869-x.
Details
Researchers at the Tokyo Metropolitan Institute of Medical Science・Laboratory of Protein Metabolism team developed a new method for determining the chain length of ubiquitylated substrates.
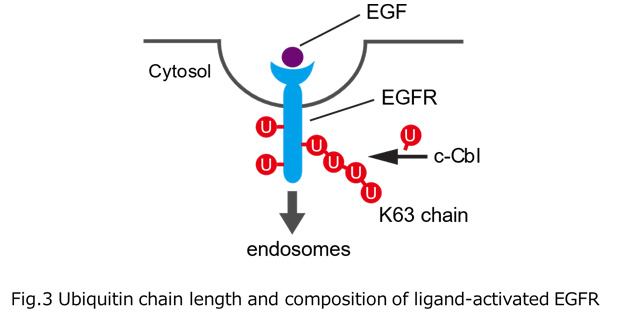
Ubiquitylation, the post-translational modification of proteins with ubiquitin, regulates almost all processes in eukaryotic cells. Because ubiquitin itself can be ubiquitylated at seven lysines and the first methionine residue, it can form polymers with distinct linkage types. Although chain length is an important parameter determining ubiquitin function, the lack of a comprehensive method for measuring this property has limited our knowledge of its physiological relevance and regulatory mechanisms. In this study, we developed a method for determining the chain length of ubiquitylated substrates. We named this method, which combines a high-affinity probe for ubiquitin and partial trypsin digestion of ubiquitin chains, ‘ubiquitin chain protection from trypsinization (Ub-ProT)’(Fig.1). Using this method, we determined for the first time the global Ub topologies in endogenous ubiquitylated substrates from soluble yeast lysates. As a result, we revealed that the lengths of the substrate-attached Ub chains were roughly in the monomer to heptamer range. By combining the new method (Ub-ProT) with MS-based Ub quantitation (Ub-AQUA/PRM), we revealed the ubiquitin linkages involved could be divided into two groups: K6, K29, and K48 linkages were involved in longer chains, whereas K11 and K63 linkages were mainly involved in dimeric chains. To our surprise, the maximum chain lengths were only slightly altered by proteasome inhibition. By contrast, inactivation of the ubiquitin-selective chaperone Cdc48 caused a significant accumulation of elongated Ub chains, suggesting that Cdc48 antagonizes the ubiquitylation machinery and terminates chain elongation by substrate extraction. (Fig. 2). Further, we determined Ub chain topology of ubiquitylated EGFR (Epidermal Growth Factor Receptor) in HeLa cells. We revealed that ligand-activated EGFR is primarily modified with K63-linked tetra- to hexa-ubiquitins (Fig.3).
Fig.1
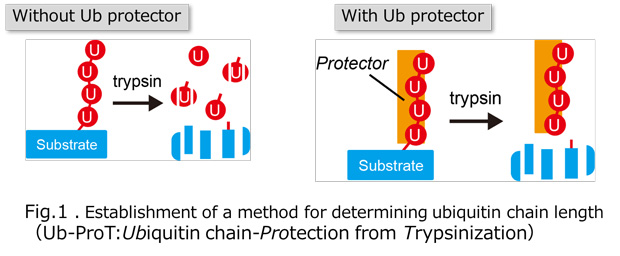
Fig.2
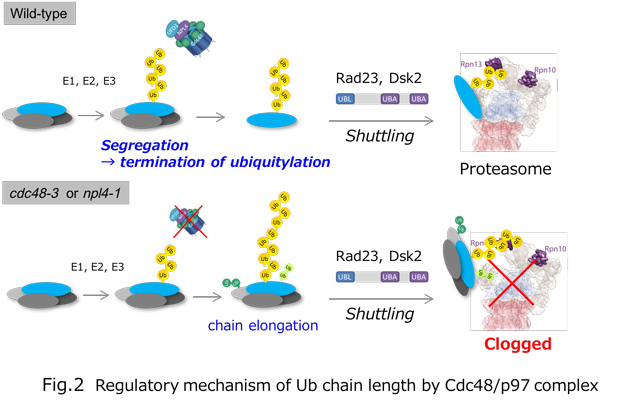
Fig.3