Dr. Yutaka Kanoh (Genome Dynamics Project), his colleagues and Geneviève Thon’s group at the University of Copenhagen published an article entitled “The fork protection complex promotes parental histone recycling and epigenetic memory” in Cell.

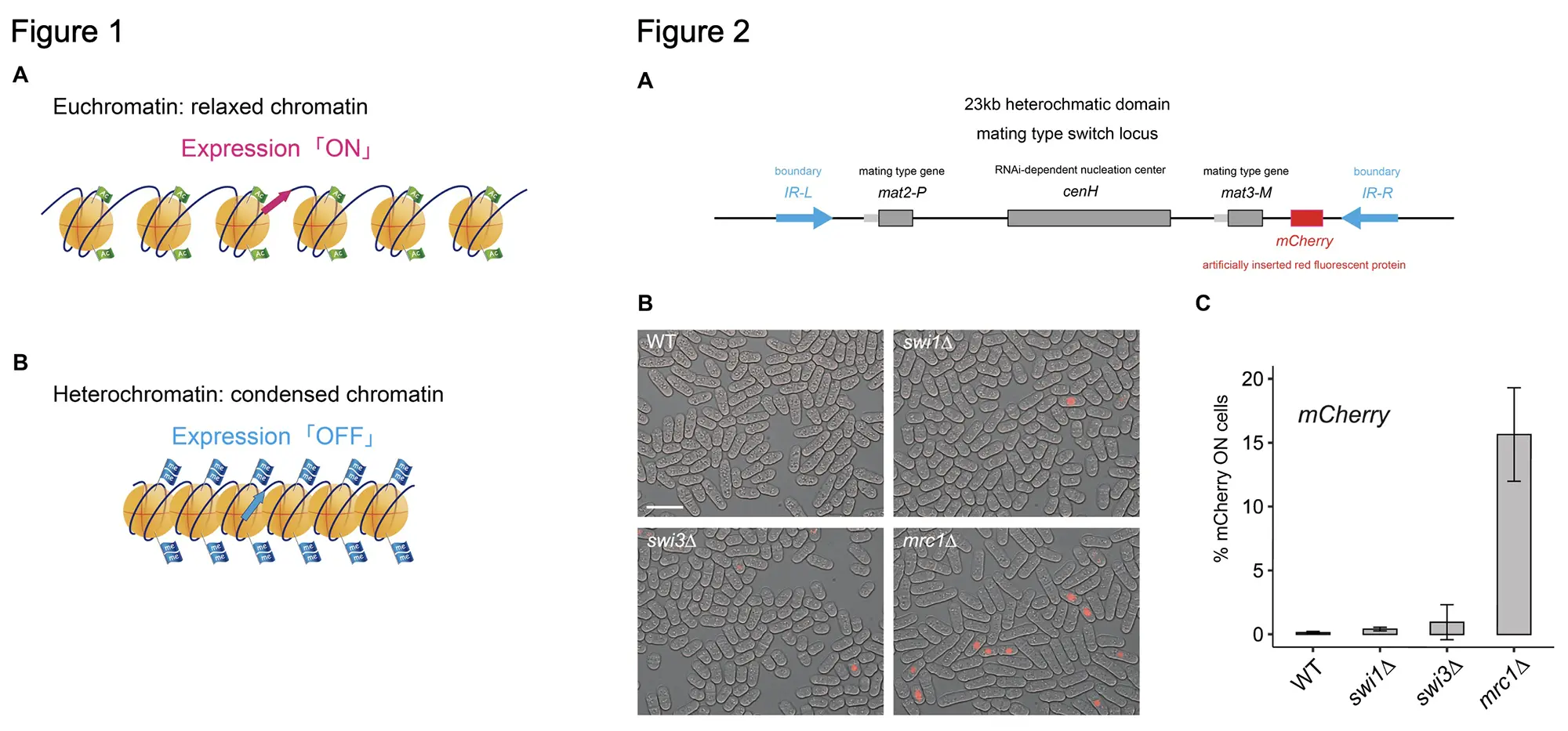
Chromatin contains the genetic and epigenetic information*1. The chemical modifications (methylation or acetylation) of histones provide the typical epigenetic information (Figure 1). The epigenetic information needs to be correctly transmitted to the next generation during DNA replication. If the inheritance does not work, the expression pattern in the daughter cells will be different from that of the parent cells and the cellular phenotypes may change. For example, in liver cells, genes responsible for liver functions are activated, but if the epigenetic information is not passed on to daughter cells, the cells may lose their characterics as the liver cells.
Our bodies are made up of many different types of cells that proliferate by repeating cell division every day. In this process, genomic DNA must first be replicated correctly. The DNA replication fork machinery consists of many proteins that are responsible for duplication of genomic DNA. The fork protection complex (FPC), composed of Mrc1-Swi1-Swi3 at the front of the replication fork machinery, promotes DNA replication and also acts as a brake when DNA damage and DNA replication stresses interfere with DNA replication fork progression, preventing the collapse of the DNA replication machinery.
Mrc1 is evolutionarily conserved from yeast to human and it transduces the signals, as a mediator of the DNA replication checkpoint, from the checkpoint sensor kinase Rad3 to the transducer kinase Cds1, which activates the DNA replication checkpoint. The HBS-deficient (∆HBS) mutant of mrc1 can bypass the Hsk1 kinase*2 mutation in a checkpoint-independent manner, although the replication checkpoint is normal. We also found that the brake on replication initiation does not work in the mrc1∆HBS cells, and the firing of the replication origin occurs prematurely. In this study, we have reported that the mrc1-null cells fails to transmit the silencing state of the mating-type switch locus, and mrc1∆HBS mutation exhibits the identical phenotype.
In S. Pombe, the mating type switch locus forms a heterochromatin structure*3 (Figure 2A), and the transcription of the genes inserted in this locus is repressed, and the gene silencing state is maintained by the heterochromatin structure. We have revealed that the silencing in the locus was frequently derepressed in the strain lacking mrc1and in the mrc1∆HBS strain (Figure 2B). This evidence indicates that the HBS domain (782-879 amino acids), to which Hsk1 kinase binds, is essential for the inheritance of epigenetic information required for gene silencing.
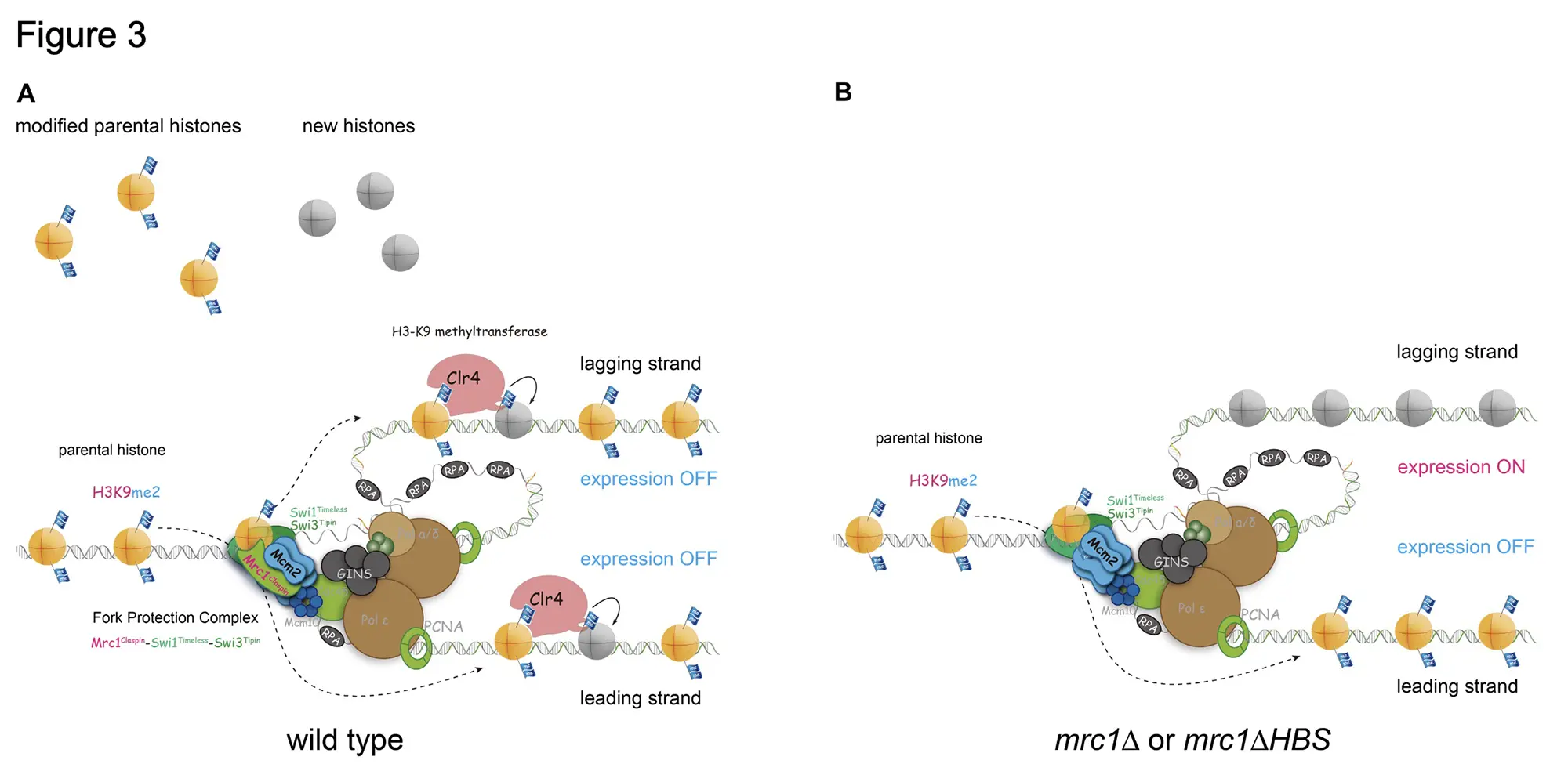
Using the SCAR-seq*4 technique, we examined whether the parental histones were redistributed on the daughter chromosome. In the wild type strain, the methylated parental histones H3-H4 were redistributed symmetrically on both the leading and lagging strands. In the mrc1∆HBS strain, the methylated parental histones were accumulated on the leading strands. Thus, it was shown that Mrc1 is essential for the symmetric distribution of epigenetic information to daughter cells (Figure 3A&B).
Previously, it was reported that Mcm2, a component of DNA replication helicase that unwinds the double strand DNA, is also required for the symmetric distribution of the parental histone to daughter chromosome, especially lagging strands (Figure 3). We predicted the structural interaction of Mrc1, Mcm2 and H3-H4 histone using AlphaFold2*5, which performs the prediction by an artificial intelligence (AI) from an accumulated protein structure database. This prediction provided the possibility that the HBS domain of Mrc1 captures the histone H3-H4 interacting with Mcm2. In fact, it was demonstrated this possibility that Mrc1 binds to Mcm2 via H3-H4 histone by the experiments using the three purified components in vitro.
In this study, we demonstrated that Mrc1 plays a role in transmitting the epigenetic information to the next generation by precisely redistributing the modified parental histones to the both leading and lagging DNA strands after DNA replication. This function of Mrc1 was also conserved in the mammalian homolog Claspin. These results revealed that the fork protection complex, which stabilizes and promotes the DNA replication forks, plays a critical role in the inheritance of the epigenetic information to the next generation in cooperation with the replication helicase.
This study is the first to demonstrate that replication factors, which are involved in the progression and monitoring of DNA replication, play a critical role not only in the inheritance of genetic information but also in the inheritance of epigenetic information through histones. Defects in cellular responses to replication fork stalling are known to be directly associated with cancer development, with replication fork protection complex playing a central role. This finding suggests that these factors are involved in maintaining genomic stability, not only through the transmission of genetic information but also by mediating the inheritance of epigenetic information. This provides novel insights into the mechanisms of disease development, such as cancer, and suggests new strategies for cancer therapy.
This study revealed the role and mechanism of the fork protection complex, particularly of Mrc1/Claspin, in the process by which cells maintain and inherit epigenetic information. This mechanism is essential for the maintenance of the gene silencing on the genome.
The maintenance and inheritance of epigenetic information play crucial roles in processes such as development, cancer, and aging. Therefore, understanding this mechanism will deepen our knowledge of cell fate determination and diseases (such as cancer and developmental disorders) and could suggest new therapeutic targets, contributing to the prevention, diagnosis, and treatment of diseases.