Tokyo Metropolitan Institute of Medical Science
2-1-6 Kamikitazawa, Setagaya-ku, Tokyo 156-8506, Japan< Japanese>
| |
| to Calpain Project Home |
| topics |
| members |
| introduction |
| research themes |
| publications |
| links |
| access |
| to Igakuken top |
Research Themes~for experts |
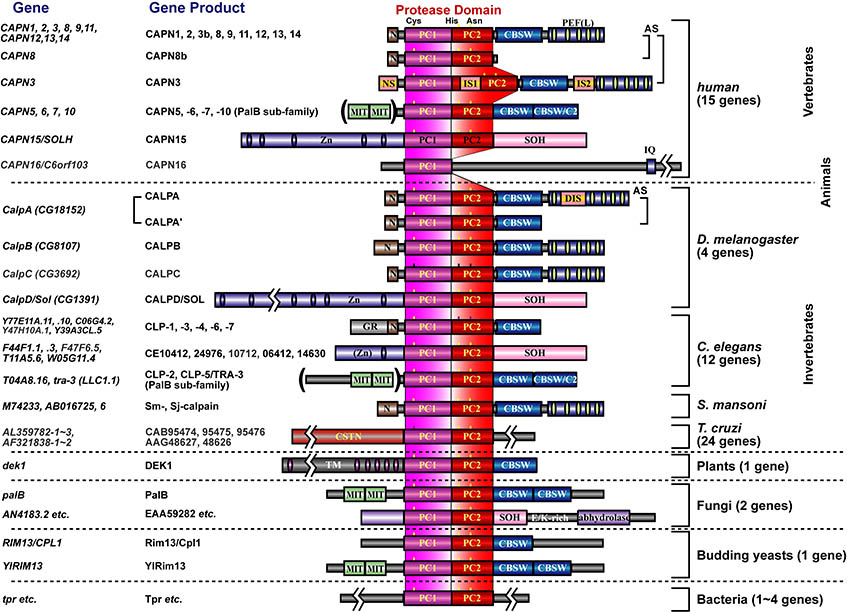
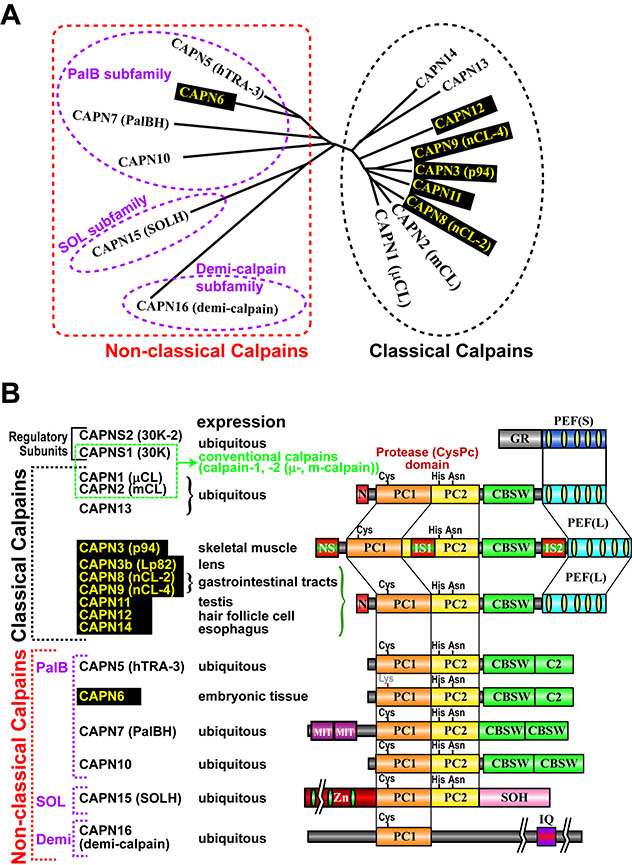
[for non-experts, please start here] In 1988, the project leader started his carrer studying calpains in late Prof. Koichi Suzuki's lab at Rinshoken. In 2004, he became independent to have his lab in Rinshoken, and, the next year, the lab's name was changed to "Calpain Project". Calpains are Ca2+-dependent intracellular processing proteases that limitedly proteolyze substrate proteins. Laboratory of late Prof. Koichi Suzuki had determined the complete primary structure of calpain for the first time in the world, and shown that calpains are a "bio-modulator" involved in Ca2+-signaling. Well-studied calpains are ubiquitously expressed calpain-1 (also called μ-calpain, μCANP, or calpain-I) and calpain-2 (m-calpain, mCANP, or calpain-II), and are called "conventional calpains". They play essential and basic roles on cellular functions. In other words, calpains are "modulator proteases", which modulate structures, functions, and activities of substrates directly by proteolytic processing. Calpains have been reported to be involved in variety of biological phenomena such as memory formation, cell cycles, tumorigenesis, cerebral and myocardial diseases, and diabetes. In 1989, we found a novel mammalian calpain catalytic subunit similar to (but distinct from) catalytic subunits of calpain-1 (CAPN1, also called μCL or μ80K) and calpain-2 (CAPN2, also called mCL or m80K). We named it "p94" after its molecular mass, but the name was changed several times (called calpain-3, or nCL-1), and finally fixed to be "CAPN3". CAPN3, different from CAPN1 or CAPN2, showed predominant expression in skeletal muscles. Four years after, we found other "tissue-specific" calpains, CAPN8 (also called nCL-2) and CAPN9 (also called nCL-4) as well as other "ubiquitous" calpains. As a result, we showed that calpains constitute a superfamily (Fig. 1), and the calpain research field was outspread to all living organisms from humans to yeasts. Tissue-specific calpains are closely related to the functions of tissues they express, and thus enable us to study calpains in the physiological contexts. Therefore, our studies mainly focus on tissue-specific calpains.
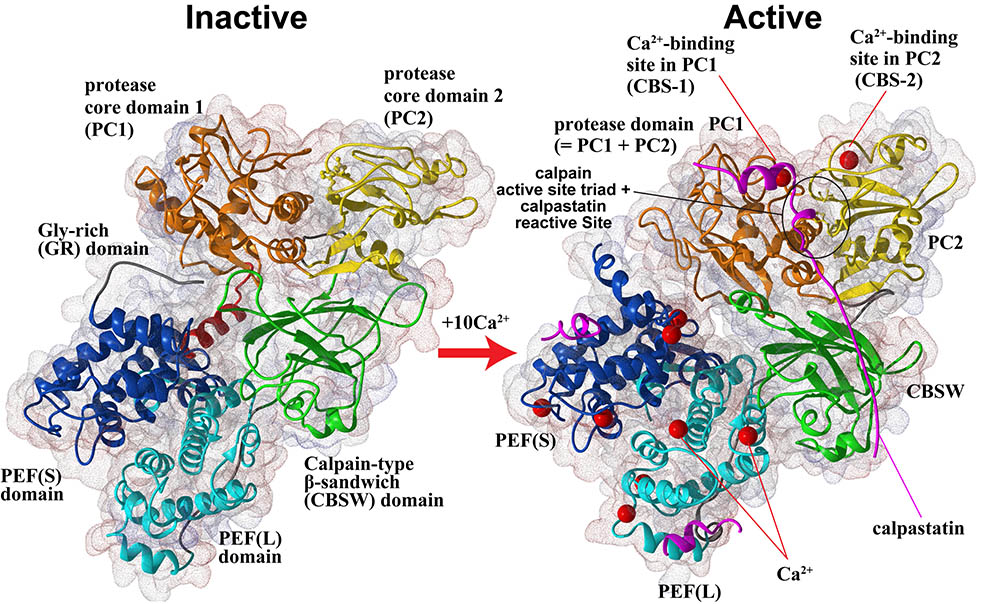
Furthermore, in collaboration with Prof. Walfram Bode's group, we have solved the 3D structure of inactive (Ca2+-unbound) calpain-2 (Fig. 2, left), and together with other information on calpain structures, structure-function relationship are being studied in our lab.
Humans have 15 calpain genes, representative products of which are shown in Fig. 3. Calpains other than CAPN1 and CAPN2 are called "unconventional calpains". As shown in Figs. 1 and 3, CAPN1 and CAPN2 consist of a well-conserved protease domain called "CysPc", which is divided into two protease core domains, PC1 and PC2, a calpain-type beta-sandwich (CBSW) domain, and penta-EF-hand (PEF) domain. CAPN3, 8, 9, 11, 12, 13, and 14 also have similar domain structures, and are called "classical calpains" (Fig. 3). Other calpains do not have CBSW or PEF "non-classical calpains". Aside from structural classification, calpains can be grouped into two by tissue distributions: relatively ubiquitous ones (CAPN1, 2, 5, 7, 10, 13, 14, 15, 16), and those expressed predominantly in a few tisses/organs (CAPN3, 6, 8, 9, 11, 12) (Fig. 3).
As another point of view, we also focus on a unique unconventional calpain, CAPN7 conserved from yeats to humans.
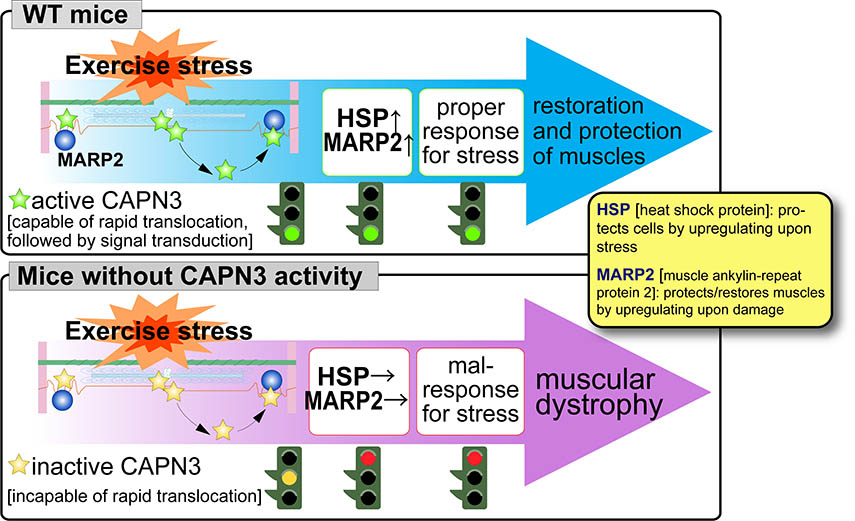
In 1993, we found a unique characteristics of CAPN3, i.e., disappearance in less than 10 min by rapid and exhaustive autolysis just after translation. We also found that a gigantic elastic muscle protein, connectin/titin, interacts with CAPN3. Although physiological meaning of CPAN3-connectin/titin interactions are still elusive, this is one of the most important sutdy focuses in the calpain research field, especially in respect of muscular dystrophies as described below. In 1995, Prof. Jacques S. Beckmann's group discovered that pathogenic mutations of human CAPN3 are responsible for limb-girdle muscular dystrophy type 2A (LGMD2A, also called "calpainopathy"). This finding clearly showed that CAPN3 is essential for skeletal muscle functions, and, since then, studies on CAPN3 has been developing in relation with LGMD2A pathogenic mechanisms. We use biochemical and genetic analyses on mice genetically modified in calpain-related genes (transgenic, knock-out, and protease-inactive (active site Cys129 changed to Ser [CAPN3:C129S]) knock-in mice) to elucidate physiological and patho-physiological functions of CAPN3. Using CAPN3:C129S knock-in mice, we have shown that a loss of protease activity of CAPN3 is responsible for LGMD2A. Moreover, CAPN3 dynamically changes its cellular localization its activity-dependently, and this movement is important for stress-response of skeletal muscle (Fig. 4, please see here for the detail).
Biochemical analyses partially clarified enzymological characteristics of CAPN3, and we have established an assay method for CAPN3 activity utilizing yeast genetics (p94-trapping), and showed that CAPN3 is activated not only by Ca2+, but also by Na+. CAPN3 is the first example of Na+-dependent intracellular enzyme (please see here for the detail).
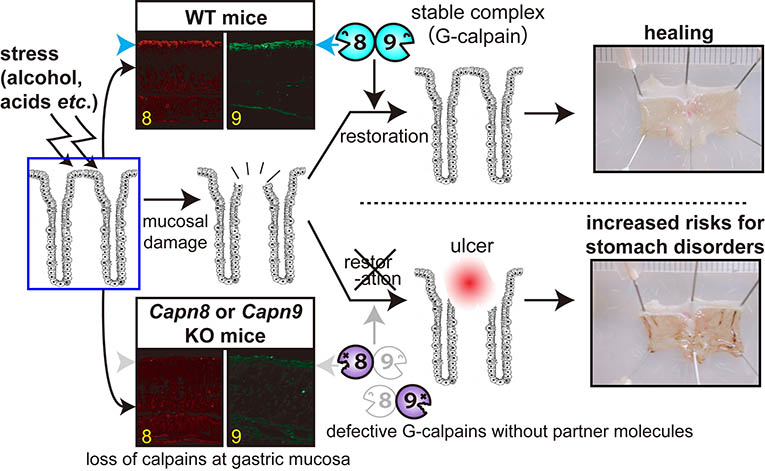
Studies using knock-out and inactive knowk-in mice of CAPN8 and CAPN9 shwed that both calpains are required for stomach mucosal defence mechanisms against stress. Loss of either CAPN8 or 9 leads to significant stomach ulcer upon alcoholic stress. Furthermore, CAPN8 and CAPN9 form 1:1 complex, and function together in the stomach. We designated it G-calpain (G: gastric) (Fig. 5, please see here for the detail).
Single nucleotide polymorphisms (SNPs) reported for human CAPN8 and CAPN9 includes non-sense mutations and mis-sense mutations that changes amino acid residues highly conserved among calpain family members. These must perturbe functions of CAPN8 and CAPN9. Actually, our in vitro assay showed that these mutant CAPN8 and CAPN9 did not have activity. [3] Other calpains, CAPN6, CAPN7, CAPN11, CAPN12 Other tissue-specific calpains, CAPN6, CAPN11, and CAPN12, are also expected to be involved in tissue-specific functions. For an example, CAPN6 was shown to be involved in supression of skeletal muscle development. CAPN6 has Lys at the position of the conserved active site Cys, indicating no proteolytic activity with this molecule. CAPN6 is shown to be involved in regulation of microtubules and RNA splicing, but it is so far unknown how these molecular functions connect with observed phenotype of its knock-out mice, i.e., significantly larger skeletal muscle (and thus body) development than WT mice. CAPN11, 12 and 14 are predominatnly expressed in testis, skin, and esophagus, respectively, and their specific functions in these organs are a center of attention. Indeed, recently CAPN14 has been shown to be responsible for eosinophilic esophagitis. [Perspective] It's been more than a quater century since we found CAPN3, and also benn more than 20 years since CAPN3 was reported as a gene responsible for LGMD2A, Studies on CAPN3 have been hampered mainly due to its unique strong auto-degrading activity. However, utilizing genetics, proteomics, and bioinformatics, we want to clarify structure-function relationships of CAPN3 and molecular pathogenic mechanisms of LGMD2A. Using genetically modified mice, we have shown that physiological functions of G-calpain or CAPN8/9 are related to stomach stress response and gastric ulcer; however, its molecular mechanisms are still elusive, which we want to elucidate by further analyses of our mutant mice. Finally, together with information about other calpains, we want to reveal the whole landscape of the calpain system, which sustains the bases for cellular functions as a molecular network. |
Contact
Calpain Group, Department of Basic Medical Sciences,
Tokyo Metropolitan Institute of Medical Sciences
Shoji Hata, Ph.D., Group Leader
2-1-6 Kamikitazawa, Setagaya-ku, Tokyo 156-8506, Japan; FAX: 03-5316-3163
E-mail:
URL: http://www.igakuken.or.jp/calpain/indexEnglish.html