
- HOME
- Circadian Clock Project
Circadian Clock Project
Circadian Clock and Lifespan/Aging Timer Project
Achievements in 2024
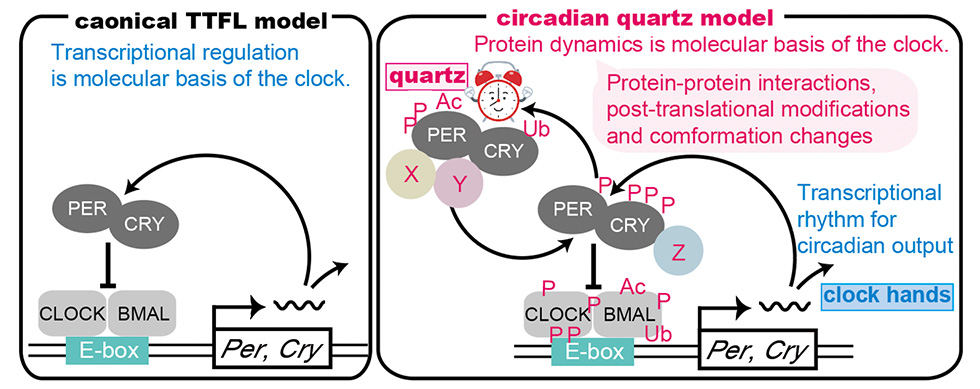
<objective 1. circadian quartz>
How does the circadian clock autonomously oscillate with a period of 24 hours? While the canonical TTFL is an important component of the clock that regulates circadian expression of downstream genes, we believe that the time-counting mechanism of the clock is regulated by protein dynamics, which includes protein-protein interactions, post-translational modifications and conformational changes of clock proteins. Thus, TTFL is required for clock read-out and is akin to the hands of the clock, while protein dynamics may be more similar to the quartz timer in the clock. Currently we are studying TTFL-independent protein-based clock components to identify the quartz timing mechanism.
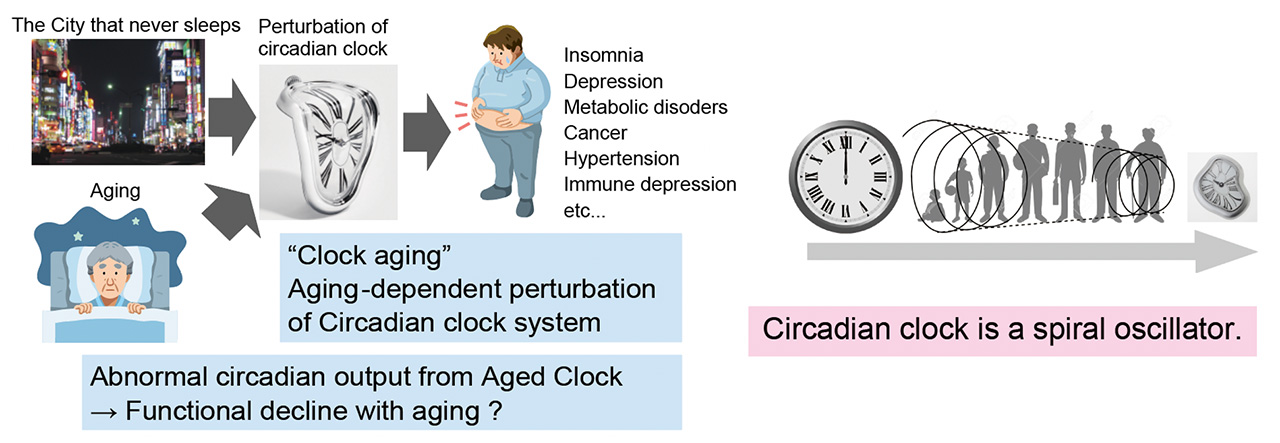
<objective 2. clock aging>
Disruption of the circadian clock causes dysregulation of gene expression rhythms. This leads to functional declines including aging-associated declines, which we refer to as “clock aging”. We are studying the molecular mechanisms of how aging disrupts the functional rhythms of the circadian clock and how clock perturbations cause aging-associated symptoms.
Publications
Papers in 2024
- Otobe et al., (2024) “Phosphorylation of DNA-binding domains of CLOCK-BMAL1 complex for PER-dependent inhibition in circadian clock of mammalian cells.” Proc. Natl. Acad. Sci. USA, 121(23): e2316858121
- Hiroki and Yoshitane (2024) “Ror homolog nhr-23 is essential for both developmental clock and circadian clock in C. elegans.” Communications Biology, 7(1):243
- Masuda et al., (2024) “TRAF7 determines circadian period through ubiquitination and degradation of DBP.” Communications Biology, 7(1):1280.
Key papers
- Terajima et al., (2017) “ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm.” Nature Genetics, 49(1): 146-151.
- Imamura et al., (2018) “ASK family kinases mediate cellular stress and redox signaling to circadian clock.” Proc. Natl. Acad. Sci. USA, 115(14): 3646-3651.
- Masuda et al., (2020) “Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch.” Proc. Natl. Acad. Sci. USA, 117(20): 10888-10896.
- Abe et al., (2022) “Rhythmic transcription of Bmal1 stabilizes the circadian timekeeping system in mammals.” Nature Communications, 13 (1): 4652.

