- HOME
- Higher Brain Function Project
Higher Brain Function Project(formerly Learning and Memory Project)
Principal of Higher Brain Functions Elucidated from Functional Algorithm of Compact Brain
Achievements in 2024
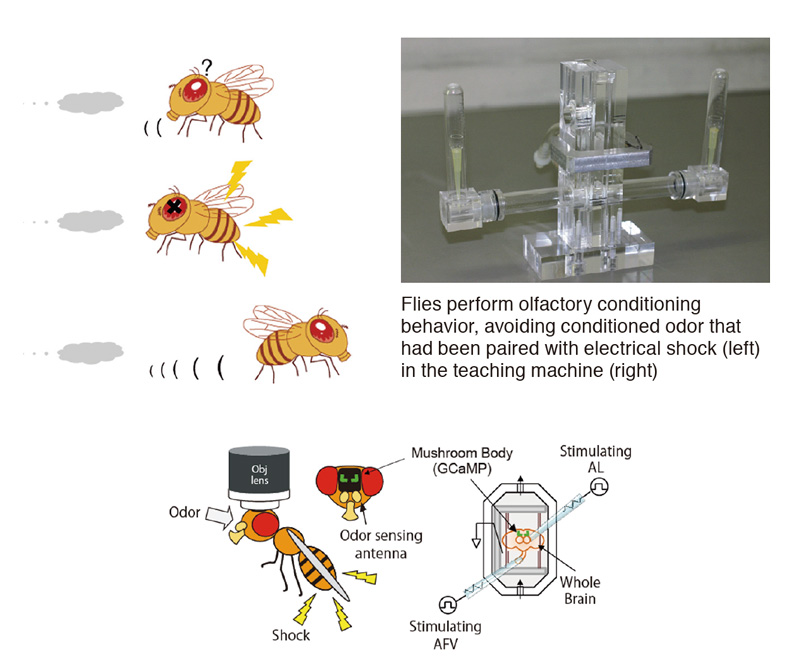
In addition to behavioral and genetic approaches, we use in vivo and ex vivo imaging techniques to characterize physiological properties of memory-associated neural networks. Our goal is to understand how the brain forms associations between specific sensory signals and positive and negative preferences, how these associations are stored in the brain in neural memory networks, and how they are later recalled at appropriate times. We further aim to understand how memory-associated genes and neuromodulatory systems regulate function of these networks.
Publications
Key papers
- Miyashita et al. (2023). "Glia transmit negative valence information during aversive learning in Drosophila" Science 382: eadf7429.
- Ueno K et al. (2020). “Carbon monoxide, a retrograde messenger generated in post synaptic mushroom body neurons evokes non-canonical dopamine release.” J Neurosci. 40, 3533-3548.
- Ueno K, et al. (2017) “Coincident postsynaptic activity gates presynaptic dopamine release to induce plasticity in Drosophila mushroom bodies.” eLife, 6: e21076.
- Hirano Y, et al. (2016) “Shifting transcriptional machinery is required for long-term memory maintenance and modification in Drosophila mushroom bodies.” Nat. Commun.7: 13471.
- Matsuno M, et al. (2015) “Long-term memory formation in Drosophila requires training- dependent glial transcription.” J. Neurosci. 35: 5557-5565.
- Yamazaki D, et al. (2014) “Glial dysfunction causes age-related memory impairment in Drosophila.” Neuron 84: 753-763.
- Hirano Y, et al. (2013) “Fasting Launches CRTC to Facilitate Long-term Memory Formation in Drosophila.” Science 339: 443-446.
- Miyashita T, et al. (2012) “Mg2+ block of Drosophila NMDA receptors is required for long- term memory formation and CREB-dependent gene expression.” Neuron 74: 887-898.